Check our deep learning paper on spot detection in microscopy data here
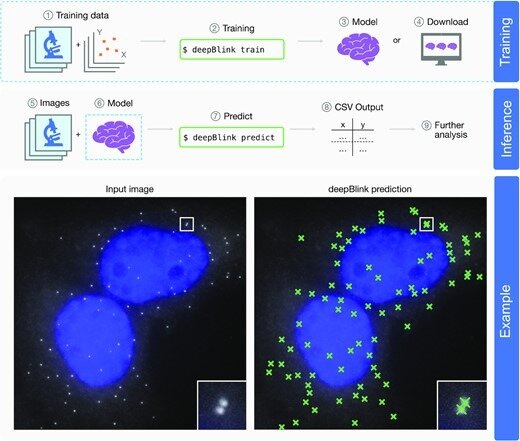
Overview of deepBlink’s functionality. deepBlink requires a pre-trained model that can be obtained by training from scratch using custom images and coordinate labels (1–3) or downloaded directly (4). To predict on new data, deepBlink takes in raw microscopy images (5) and the aforementioned pre-trained model (6) to predict (7) spot coordinates. The output is saved as a CSV file (8) which can easily be used in further analysis workflows (9). An example use case is shown for a smFISH analysis with blue indicating DAPI staining.