In this work, led by Mattia Ubertini and Nessim Louafi, we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication. Check the full manuscript out on bioRxiv! https://www.biorxiv.org/content/10.1101/2025.09.24.678119v1
New member
Warm welcome to Jonas Oefelein, who joins the lab as a PhD student!
New preprint online: Enhancer control of promoter activity and variability via frequency modulation of clustered transcriptional bursts
Find our new preprint online: https://www.biorxiv.org content/10.1101/2025.03.26.645410v1.full
Our new study challenges existing models of mammalian promoter operation, and reveals that enhancer position within a cis-regulatory landscape critically controls the timing and variability of transcriptional output in single cells.
Welcome to a new undergraduate student
This May we welcome Iris Jehel, who joins the lab for 4 month internship!
End of 2024
Last week we had several team-building events : the traditional end-of-year FMI party, then we went out for Christmas lunch and ice-skating with the lab, and finally the Novartis after-work event on campus. 2024 was another successful and productive year, and we are excited for what 2025 will bring! Happy holidays season and successful New Year!
Two EMBO Postdoc Fellowships in the lab
The end of 2024 brings fantastic news! Congratulations to our two postdocs, Verena and Mattia, who have both been awarded EMBO Fellowships! The EMBO Postdoctoral Fellowships support outstanding researchers across Europe and beyond, providing up to two years of funding to advance their scientific careers. What an exciting way to wrap up the year!
Pia Mach received the Faculty of Science Prize for her PhD thesis
Congratulations to Pia Mach, our former PhD student, for receiving the Faculty of Science Prize at the Dies Academicus of the University of Basel! The prize recognizes outstanding doctoral theses and studies.
PhD defense of Jana Tünnermann
On 2nd of December Jana successfully defended her thesis entitled “Enhancer control of transcriptional activity via modulation of burst frequency”. Great work, Dr. Tünnermann and all the best for your future endeavors!
Greg wins a poster prize at the EMBL conference!
Greg and Luca have recently attended Quantitative biology to molecular mechanisms EMBL conference in Heidelberg, where Luca gave a talk and Greg presented a poster. Congratulations to Greg for the best poster prize award!
Giorgetti lab at EMBL Transcripton and chromatin 2024
People from the lab just came back from the EMBL conference on transcription and chromatin in Heidelberg, where Jana gave a talk, and Julie and Kristina presented their posters. We listened to the amazing science and received a lot of good feedback on our projects. Moreover, it was a great opportunity to network and connect with other scientists from all over the world!
Goodbye to Yomna Gharib
Today we said goodbye to our master student Yomna Gharib, it was great to have you in the lab and we wish you all the best for your next steps!
Annual lab barbecue
Last week, we enjoyed our traditional annual lab barbecue by the Rhein, bringing together all current and former members.
New members
A warm welcome to our new lab members - Yomna Gharib, master student from Université Paris Cité, who joined in April and will stay for 5 months, and a new postdoc Dr. Verena Mutzel, who joined in June!
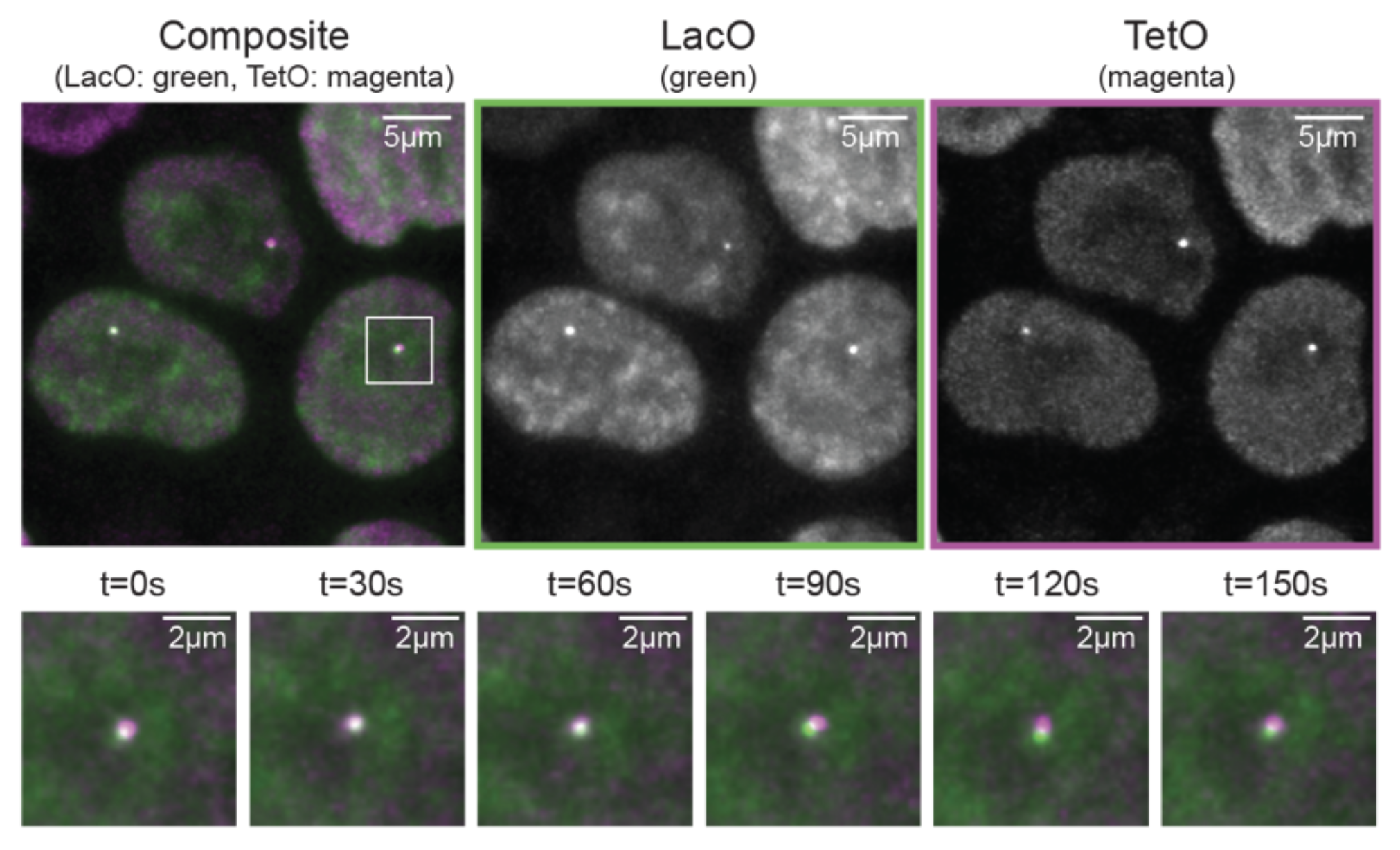
New preprint online! Live-cell imaging and physical modeling reveal control of chromosome folding dynamics by cohesin and CTCF
Our new preprint is online: https://www.biorxiv.org/content/10.1101/2022.03.03.482826v1
We imaged chromosome folding dynamics at high spatial and temporal resolution for several hours in living mESC and discovered how cohesin and CTCF control looping timing and duration!
We are looking for Master students!
We propose Master projects (min. 6 months) on these two themes:
1. To measure promoter transcription as a function of enhancer location and their mutual interaction probabilities, we recently developed a new assay enabling an enhancer to be positioned at large numbers of chromosomal sites around its target promoter (see our recent preprint). We would like a student to contribute to the development of a new version of this assay allowing to measure many enhancers and promoters in parallel.
2. Understanding enhancer-promoter communication mechanistically ultimately requires to measure how they interact in single living cells. To this end we use live-cell imaging techniques allowing us to track the position of an enhancer and a promoter in real time within the nucleus, as well as the RNA that is produced by the promoter. We are looking for a student to help with cell line engineering, microscopy and data analysis.
The student(s) working on these projects will:
- Use CRISPR/Cas9 and transposons to engineer mouse embryonic stem cells
- Perform live-cell imaging on state-of-the-art microscope systems
- Learn basic methods of quantitative imaging data analysis.
We are looking for a highly motivated and self-driven student with knowledge of basic molecular biology techniques. Experience with cell culture, microscopy and image processing is an advantage. Candidates should apply by email with a CV listing courses and internships taken during Masters studies.
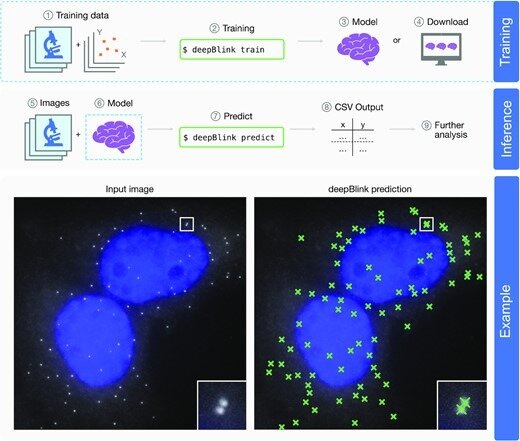
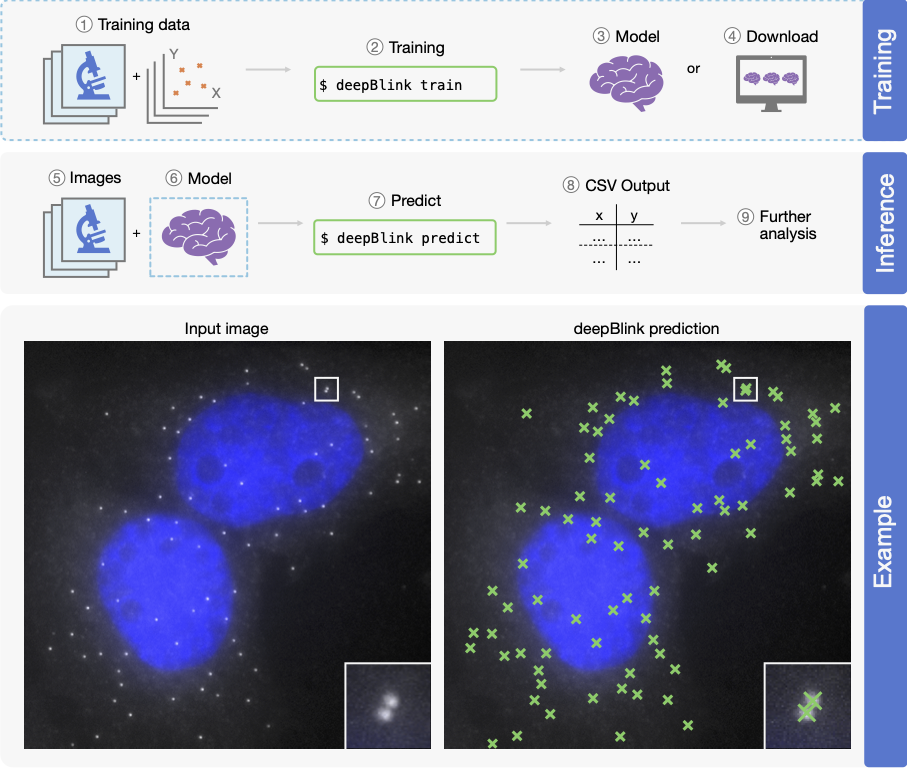
Our deep learning method to detect diffraction limited is out on Nucleic Acid Research
Check our deep learning paper on spot detection in microscopy data here
Overview of deepBlink’s functionality. deepBlink requires a pre-trained model that can be obtained by training from scratch using custom images and coordinate labels (1–3) or downloaded directly (4). To predict on new data, deepBlink takes in raw microscopy images (5) and the aforementioned pre-trained model (6) to predict (7) spot coordinates. The output is saved as a CSV file (8) which can easily be used in further analysis workflows (9). An example use case is shown for a smFISH analysis with blue indicating DAPI staining.
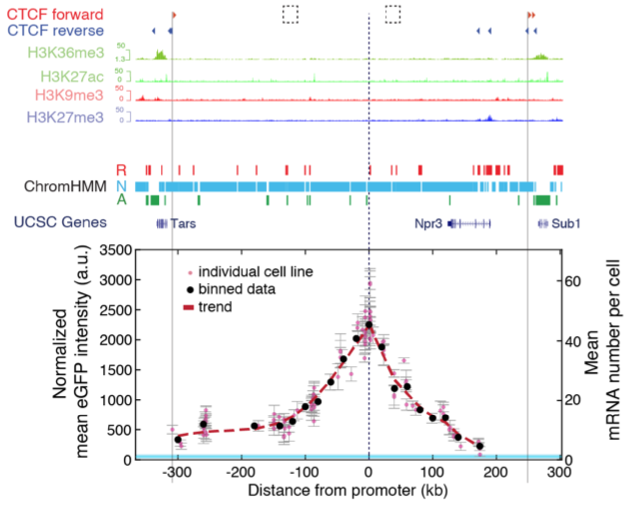
New preprint: Nonlinear control of transcription through enhancer-promoter interactions
Jessica’s and Gregory’s project is finally out on bioRxiv: https://www.biorxiv.org/content/10.1101/2021.04.22.440891v1
We use a combination of genomic engineering, quantitative measurements and mathematical modelling to address the long-standing question of how changes in enhancer-promoter genomic distance and contact frequency modulates transcription.
New preprint! A deep-learning method (deepBlink) to automatically detect and localise spots in FISH and live cell imaging data
deepBlink can detect spots in images with varying background
deepBlink does not need manual adjustment of thresholds
deepBlink outperforms benchmarking methods (TrackMate, detNet)
deepBlink is easy to use via our web interface http://deepblink.org/
The preprint of our method is out, check here https://www.biorxiv.org/content/10.1101/2020.12.14.422631v1.
The lab is growing!
Welcome to Gergely, our brand new PhD student!
Gergely has recently joined our lab to study how transcription is regulated by distal enhancers. You can read more about Gergely in the Lab members section.
We wish him a great journey with us throughout his PhD project!
Our review on 3C methods and beyond published in Molecular Cell!
The final version of our Review on methods and concepts in chromosome structure is out in Molecular Cell and can be found here.